|
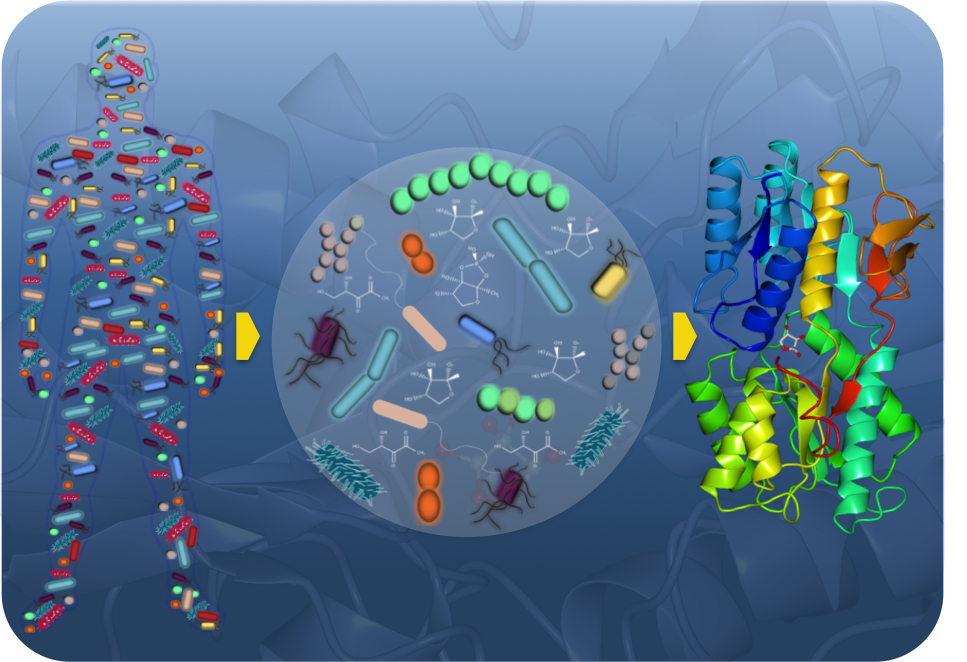
Bacteria use small chemical molecules called autoinducers to communicate with one another by a process called quorum sensing. This process enables a population of bacteria to regulate behaviours, which are only productive when many bacteria act in concert as a group, similarly to what happens with multi-cellular organisms. Behaviours regulated by quorum sensing are often crucial for successful bacterial-host relationships whether symbiotic or pathogenic. In the Bacterial Signalling laboratory, we use biochemical and genetic approaches to study the molecular mechanisms underlying quorum sensing, with an emphasis on systems promoting bacterial interspecies communication. Our research includes an integrated study involving elucidation of the chemical molecules that are used as signals, the network components involved in detecting the signals and processing information inside individual cells, and finally characterization of the behaviours regulated by these signal in multi-species bacterial consortia. Our research goal is to understand how bacterial signalling shapes multi-species bacterial microbiota communities that can be found in animals and plants and how these communities affect host physiology. In particular our group is interested in understanding how bacterial signalling plays a role in assembling, maintenance and resilience of microbiota communities. 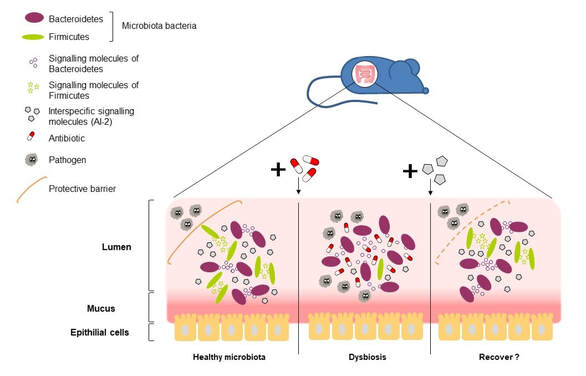
A healthy microbiota acts as a protective barrier against pathogens invasion.
Antibiotics cause disturbance in microbiota with detrimental effects on the host (dysbiosis).
Dysbiosis leads to the protective barrier destruction and susceptability to pathogens.
We are using signalling chemical molecules from bacteria communication to recover the beneficial properties of microbiota.
|
|
FUNDING